PeculiaR
We all know that floats are evil. Yet, there are moments in life when even something very well known can stab you in the back.
We all know that floats are evil. Yet, there are moments in life when even something very well known can stab you in the back.
A little background
We run all our R scripts in Docker. The interface between an R script and our environment is simple: CSV files in designated directories. This means that when a user requests that an R script (e.g. R Transformation) is run on data from table X. Our Docker component will export table X to a CSV file, then put that file inside a Docker container and then run the user R script in that container. That R script produces a CSV file Y. Our Docker component then takes this file and creates a table Y from it.
During the whole of a dull, dark, and soundless day in the autumn of the year, we received a Zendesk ticket from our customer — let’s call him Tim. He was analyzing some table from Google Analytics and then merged it back with another table from Google Analytics. His complaint was that “we changed his IDs”, because the script worked fine when he used it locally in RStudio. This is of course a very serious issue, because

It turned out, that his table was exported from Google Analytics and looked like this:
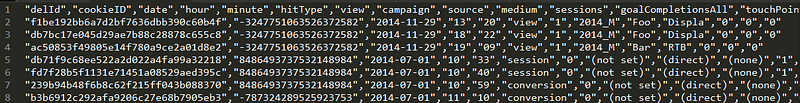
He discovered that the when the script executes (and loads data into our Storage), and attempts to join the data with another table, the values of cookieID do not match. Furthermore, he discovered that the cookieID values changed slightly e.g. from 7576593838432157975 to 7576593838432157696.
Causes
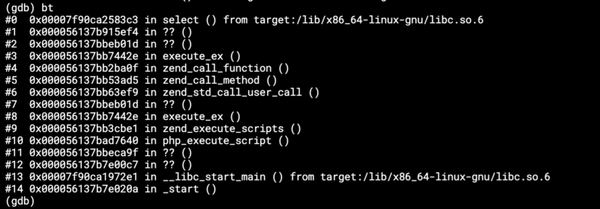
When you look at it, it’s crystal clear that it’s a floating point rounding error. But where? I pointed that out to Tim and he quickly hacked a test to check and prove that the values loaded into his R script were indeed the values returned by his R script. So his script looked like this:data <- read.table("/data/in/tables/sample.csv", sep = ",",
stringsAsFactors = FALSE, header = TRUE)
data$idRow <- 1:nrow(data) # tmp column to maintain sorting
data.backup <- data
data.backup <- data.backup %>% arrange(idRow)… snip very long script …compare1 <- paste0(data.backup$delId, data.backup$cookieID,
data.backup$date, sep = "--")
compare2 <- paste0(data_final$delId, data_final$cookieID,
data_final$date, sep = "--")
all(compare1 == compare2)
# if true - source and destination tables have same IDs
So we were a bit stuck until I realized that he missed the very first line in his test. When you load a CSV file into R, R is clever enough to guess data types for each of the columns. Unfortunately, it is stupid enough to guess data types, which cause information loss. So when you load a number like 7576593838432157975. R guesses that it is an integer, but then it realizes that it cannot actually fit it into an integer, because it uses 32bit integer which only goes up to 2147483647, even though you use a 64bit version of R. So it gets confused and casts the column to numeric, which is a floating point type and that cripples the whole thing.
You can actually check that even 64-bit R uses 32-bit integers, by calling .Machine which will give you:$double.eps
[1] 2.220446e-16$double.neg.eps
[1] 1.110223e-16$double.xmin
[1] 2.225074e-308$double.xmax
[1] 1.797693e+308… snip …$integer.max
[1] 2147483647$sizeof.long
[1] 4$sizeof.longlong
[1] 8$sizeof.longdouble
[1] 16$sizeof.pointer
[1] 8
Actually the only difference between 32-bit R and 64-bit R is in pointer size (i.e. the usable memory) and longdouble (that’s a float again):
64-bit version:$sizeof.longdouble
[1] 16$sizeof.pointer
[1] 8
32-bit version:$sizeof.longdouble
[1] 12$sizeof.pointer
[1] 4
This of course is already well documented.
Solutions
There is no way around except manually forcing a correct data type when loading the data (colClasses does the trick). In this case, there were actually no computations performed on the CookieID column, so it’s fine to cast it to character and let go. It’s noteworthy to say that a similar issue already exists on StackOverflow.
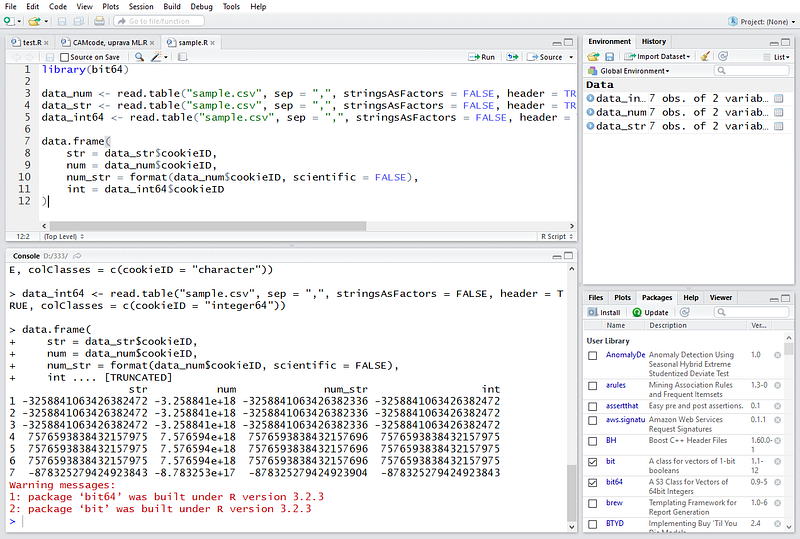
In case you do need computations, you must use 64bit integer library and also cast the column to a new type integer64 introduced with that library. You can see for yourself with the following test script:library(bit64)data_num <- read.table("sample.csv", sep = ",",
stringsAsFactors = FALSE, header = TRUE)
data_str <- read.table("sample.csv", sep = ",",
stringsAsFactors = FALSE, header = TRUE,
colClasses = c(cookieID = "character"))
data_int64 <- read.table("sample.csv", sep = ",",
stringsAsFactors = FALSE, header = TRUE,
colClasses = c(cookieID = "integer64"))data.frame(
str = data_str$cookieID,
num = data_num$cookieID,
num_str = format(data_num$cookieID, scientific = FALSE),
int = data_int64$cookieID
)
Which will give you (notice the difference between str column and num_str column which contains the numeric value converted to string):
The moral of the story (in no particular order):
- When you write tests, make sure not to skip any innocent lines
- Never trust R you can’t throw out of a window
- Don’t underestimate floating point numbers
- Strings are actually good
- Never mess with customers’ IDs
- Be careful about automatically detected things